Prominence (topography) [source wikipedia]¶
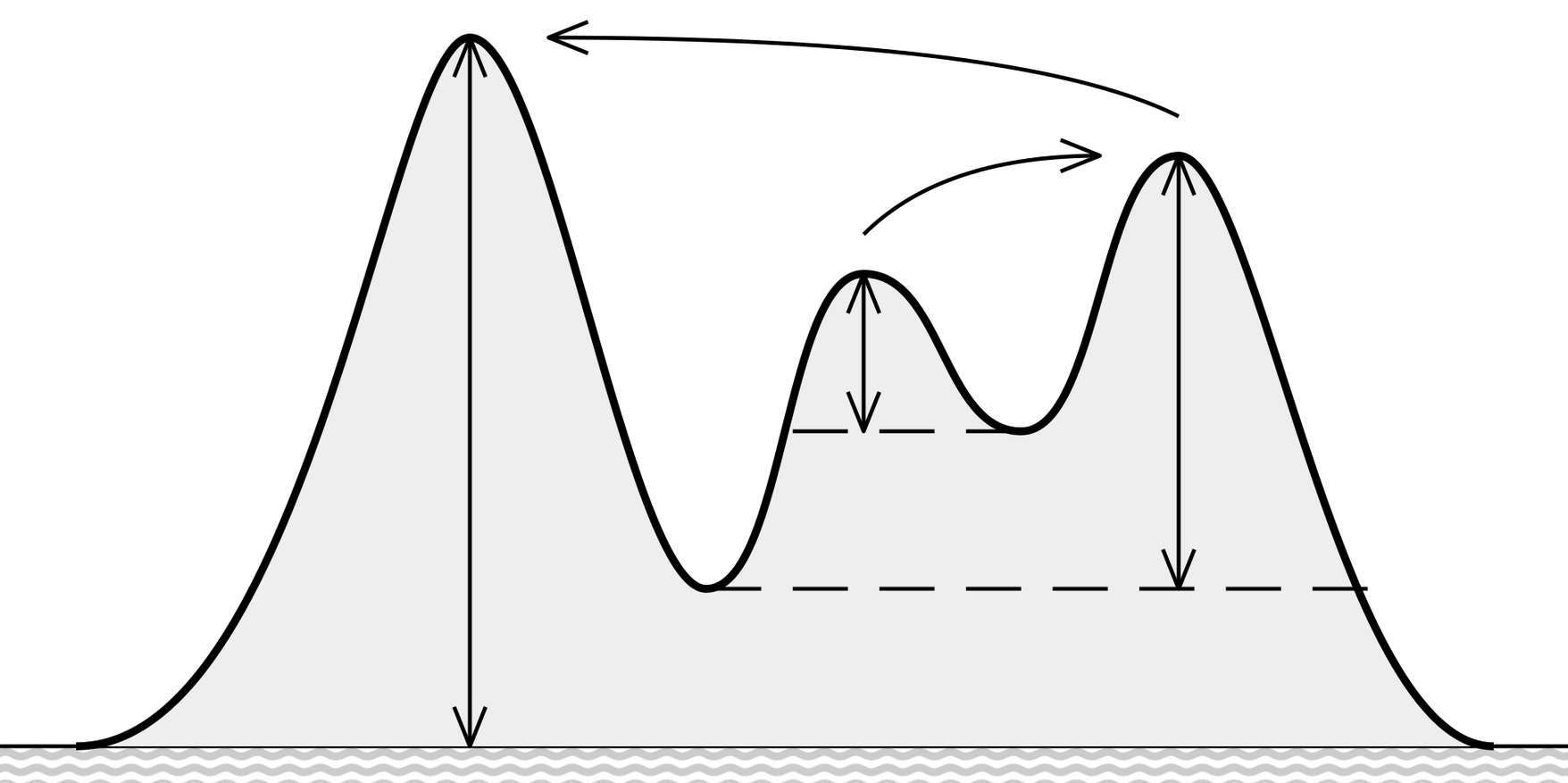
Local maximum: how low do you need to go before you can reach a higher maximum?
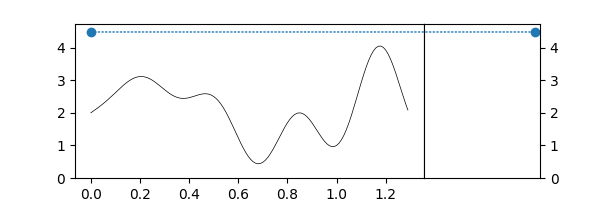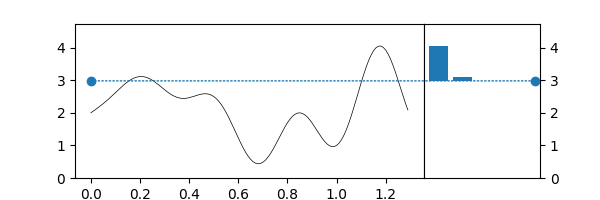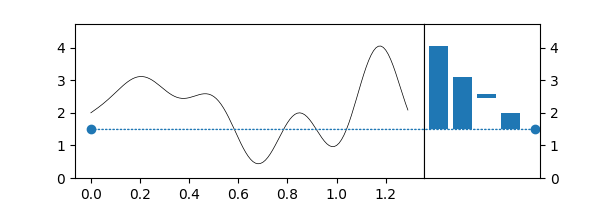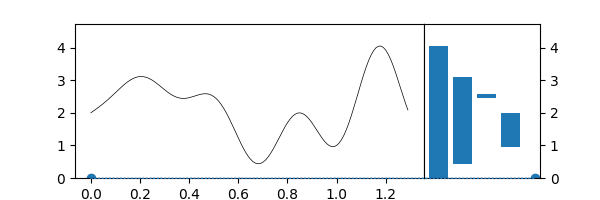
Superlevelsets¶
Superlevelsets¶
Ok, but... In topology, we compute sublevelsets¶
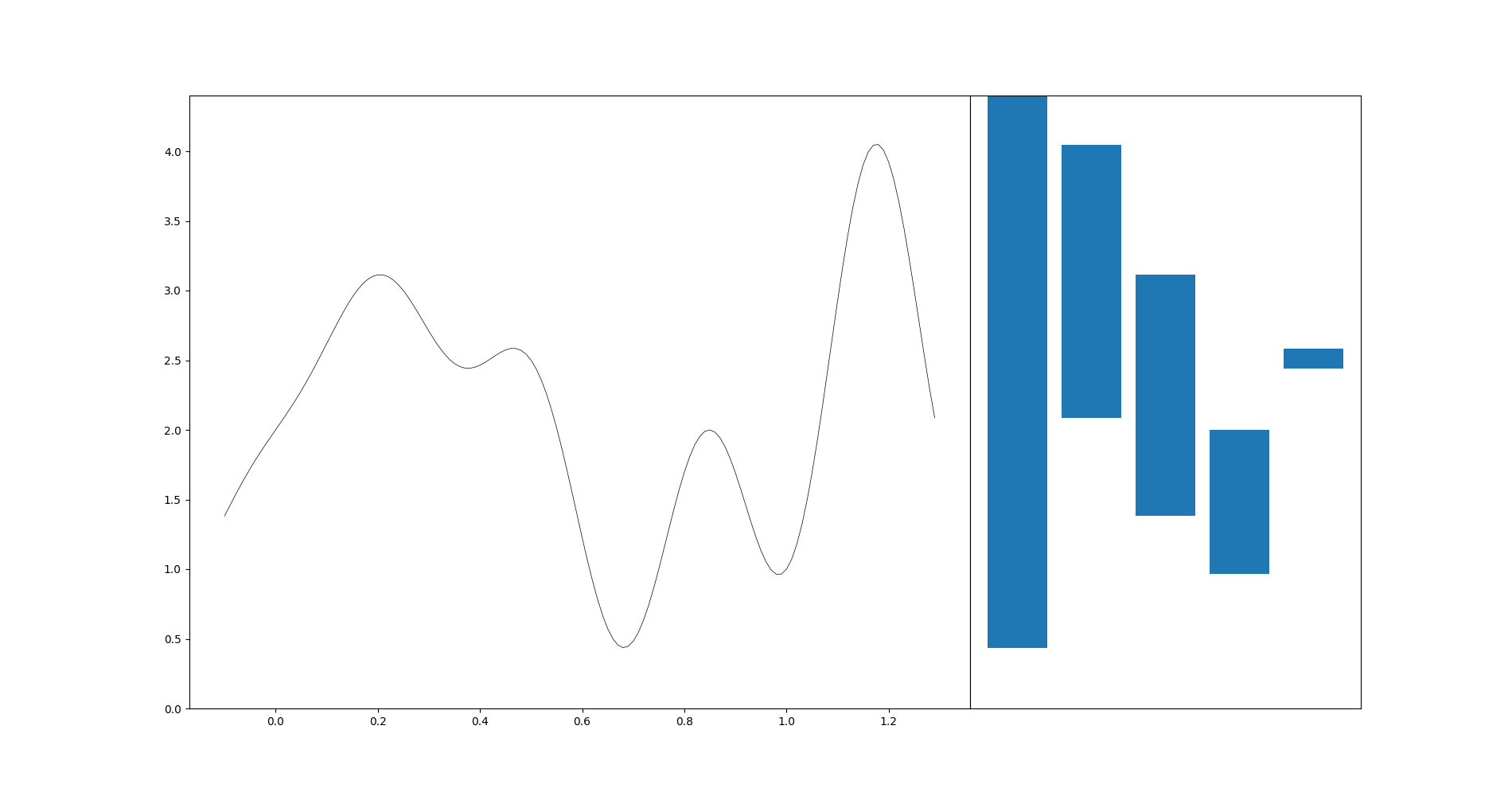
import matplotlib.pyplot as plt
import numpy as np
import gudhi as gd
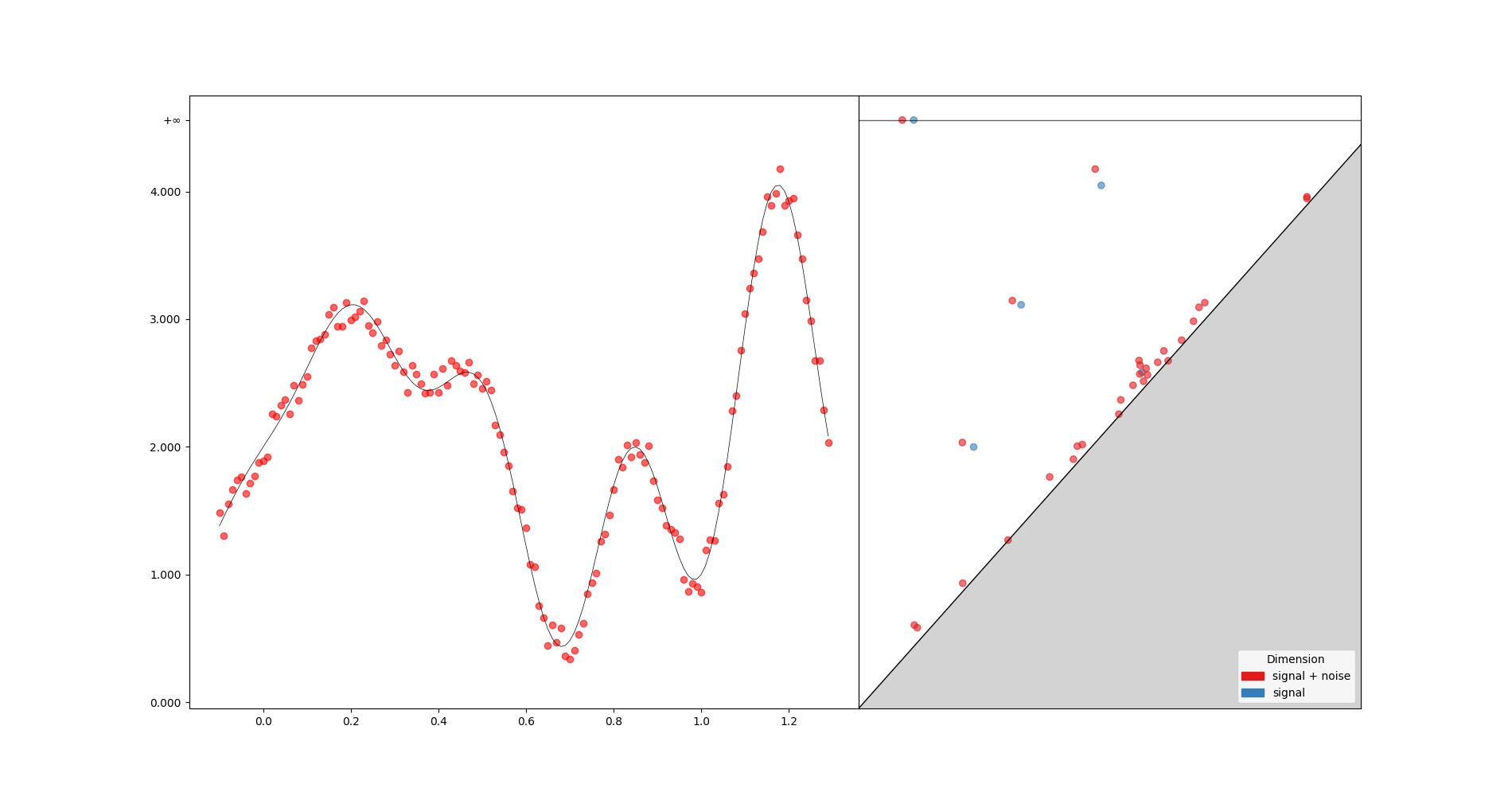
t = np.arange(-0.1, 1.3, 0.01)
s = np.sin(2 * np.pi * t) - t * np.cos(6 * np.pi * t) + 2
fig, axes = plt.subplots(ncols=2, sharey=True, figsize=(7, 1), gridspec_kw=dict(width_ratios=[4, 3], wspace=0))
fig.set_size_inches(9, 4)
axes[0].plot(t, s, "k", lw=0.5)
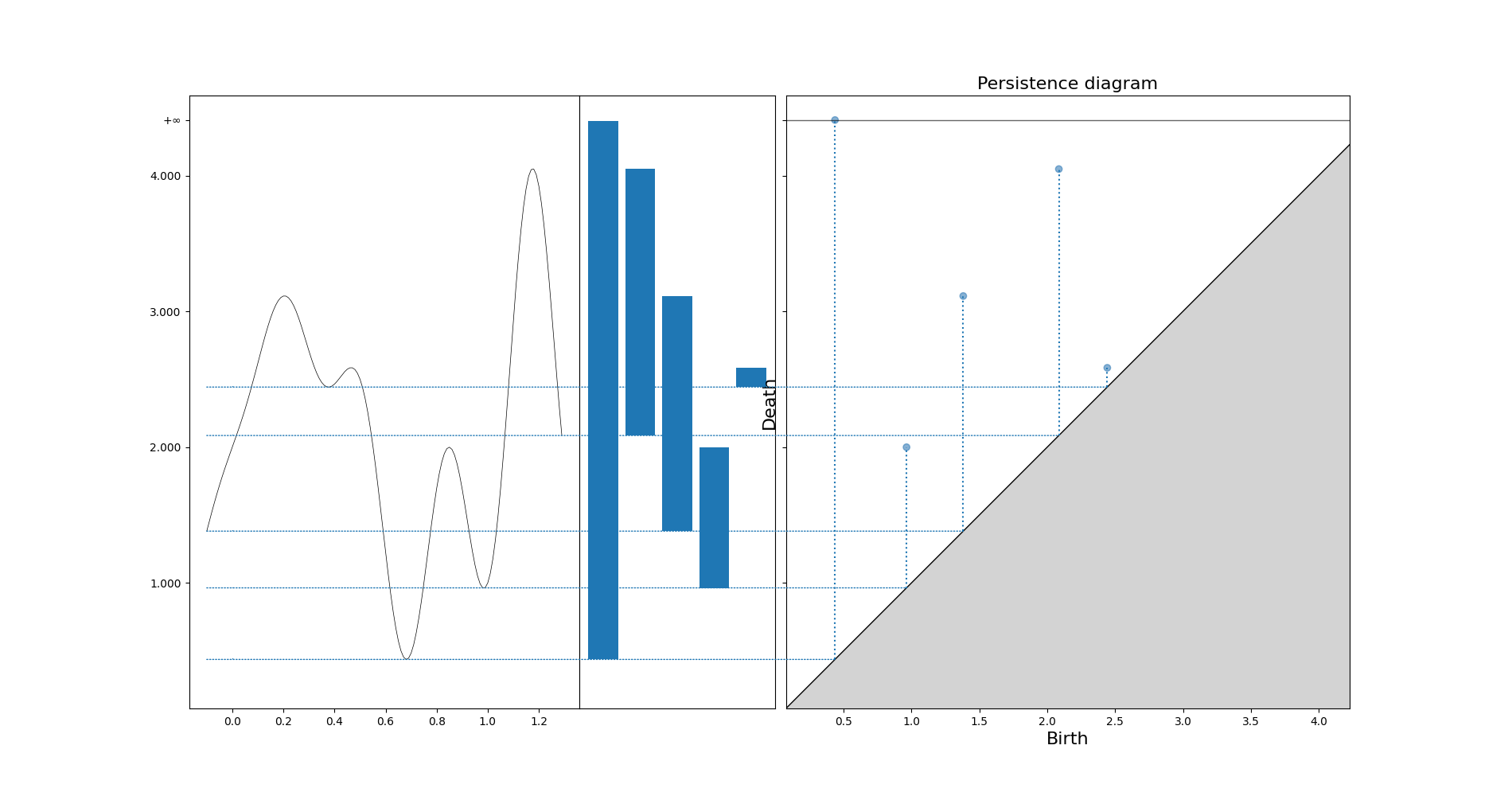
diag = gd.CubicalComplex(top_dimensional_cells=s).persistence()
_ = gd.plot_persistence_diagram(diag, legend=True, axes=axes[1])
Stability¶
\begin{equation} \lVert f - g \rVert _\infty = sup _{x \in X} \left\{ \lvert f(x) - g(x) \rvert \right\} \end{equation}
Bottleneck distance: Partial matching, the rest matched with the diagonal. The worst pair defines the cost
Stability theorem: $$ d_B ( Dgm(f) - Dgm(g) ) \le \lVert f - g \rVert _\infty $$
Representations¶
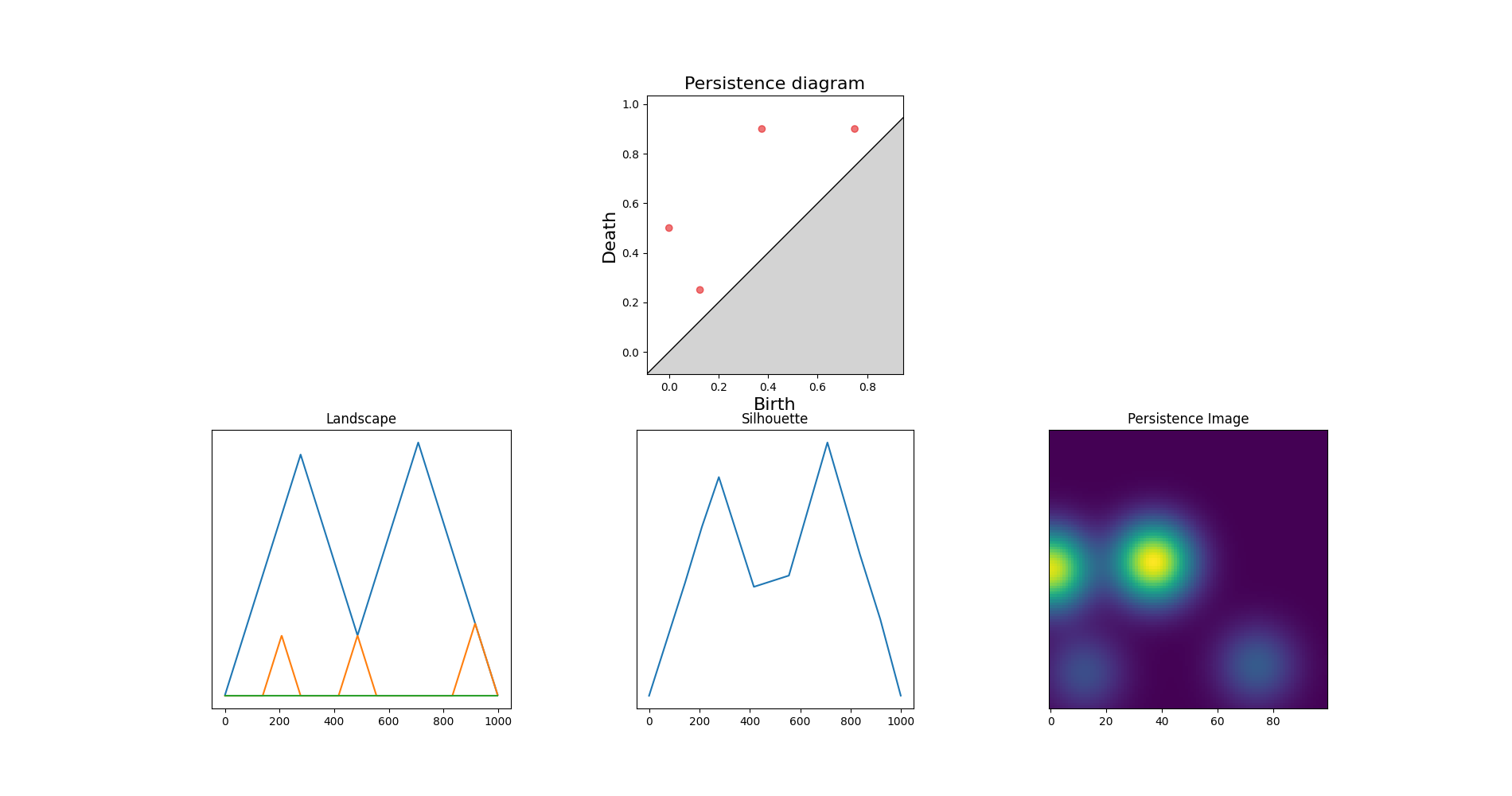
Independent (?) problem: point clouds¶
Input: point set P
Assumption: P approaches some unknown ideal object
Sublevel sets on a point cloud¶
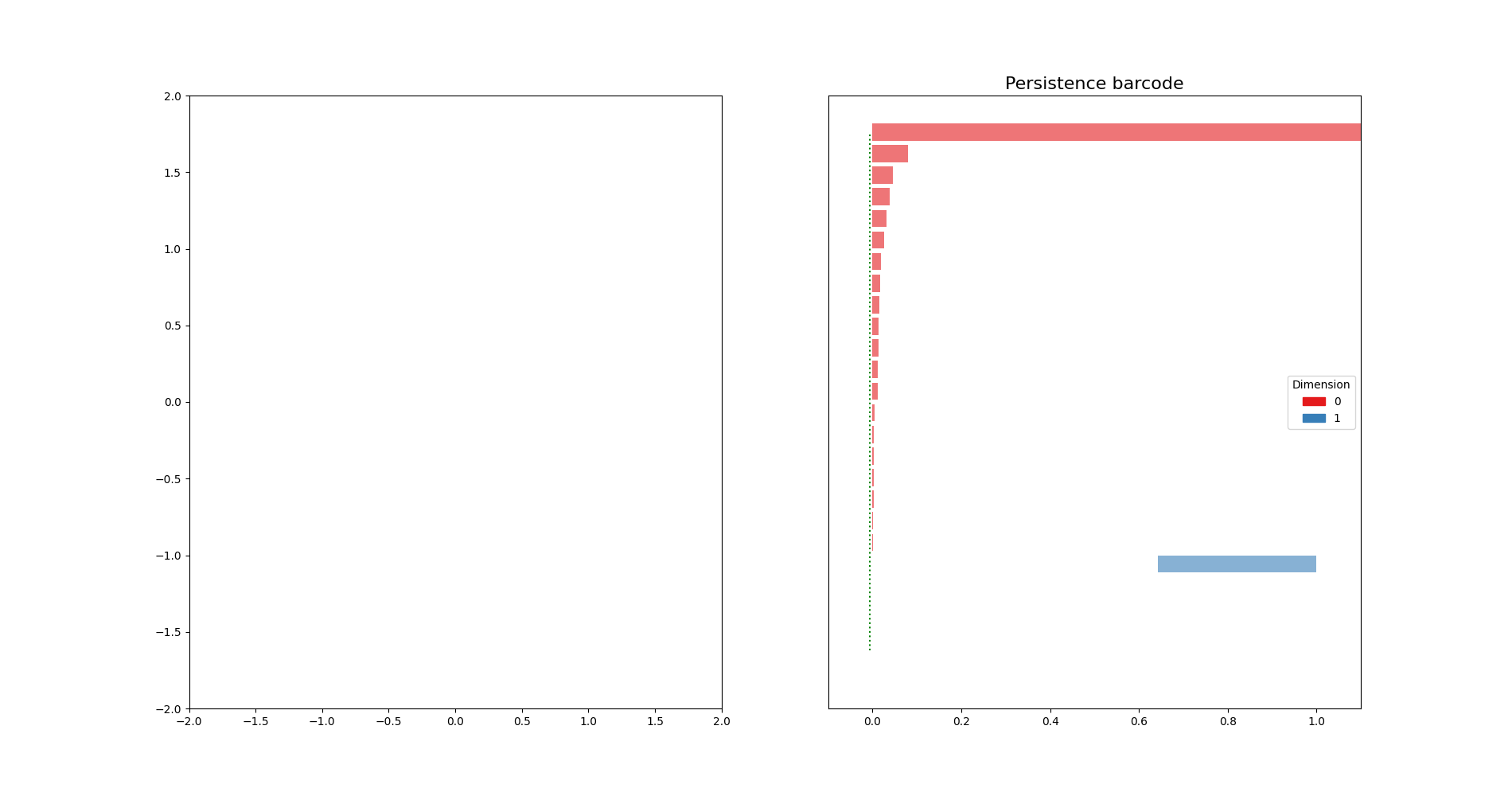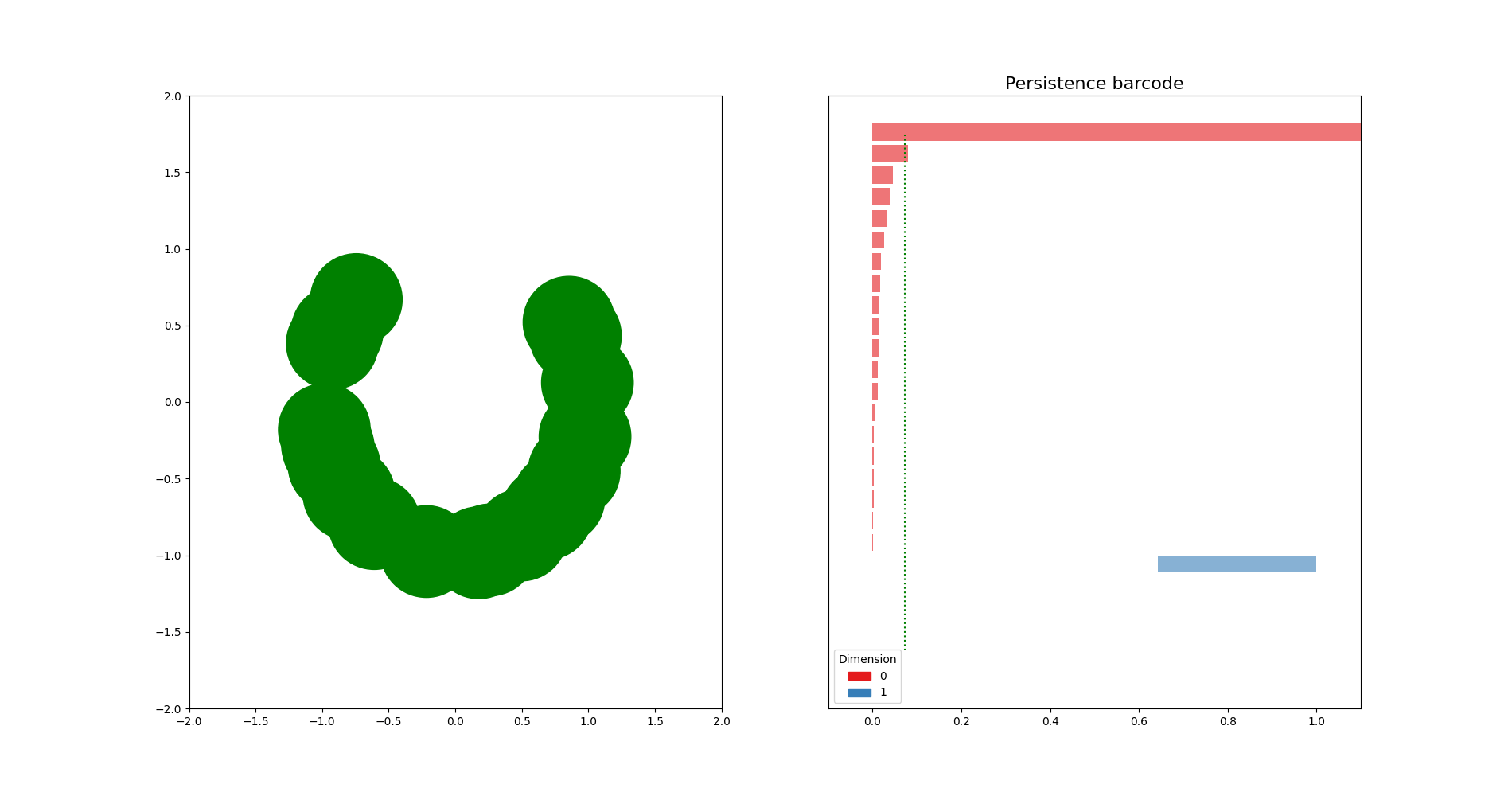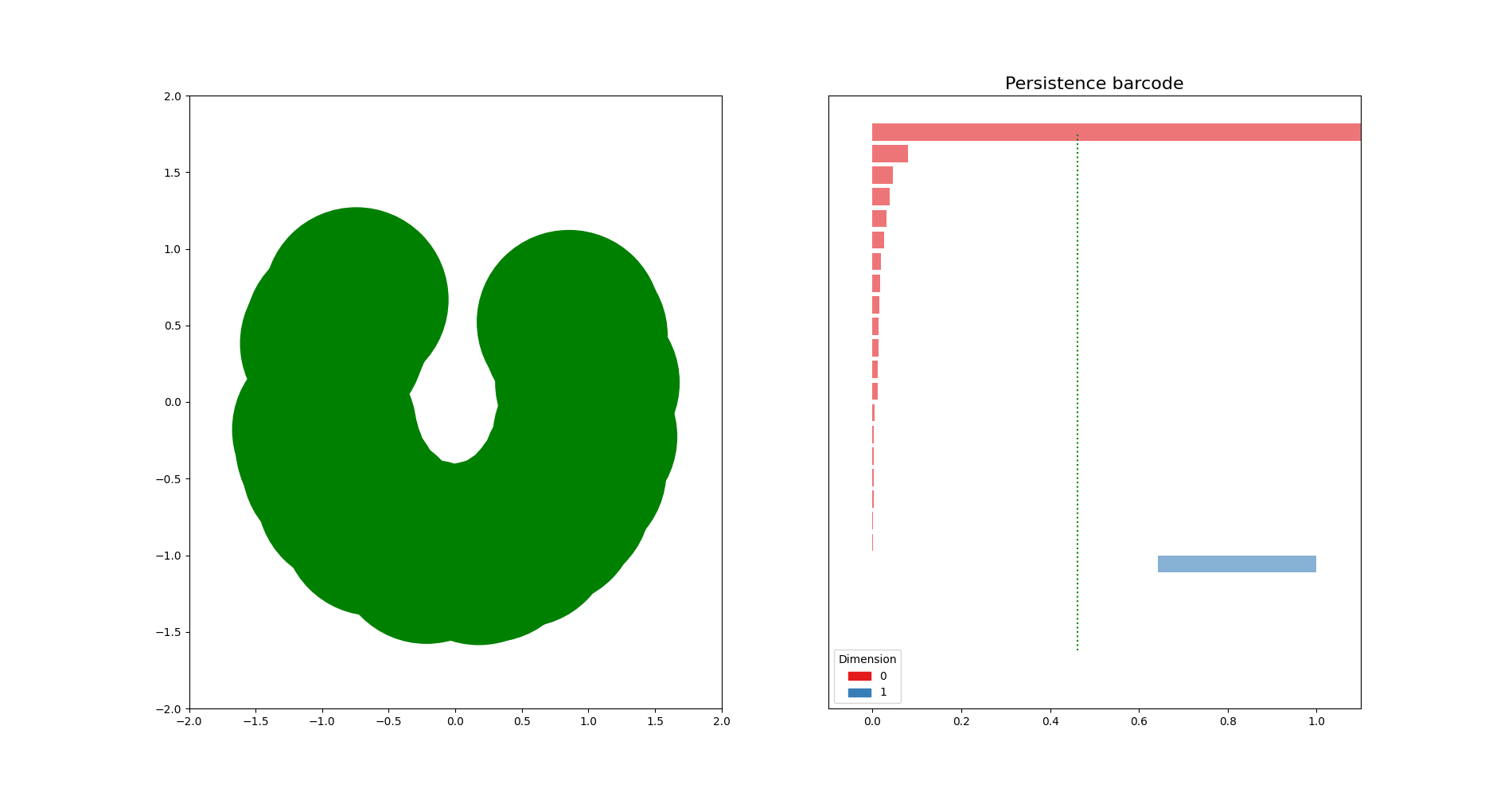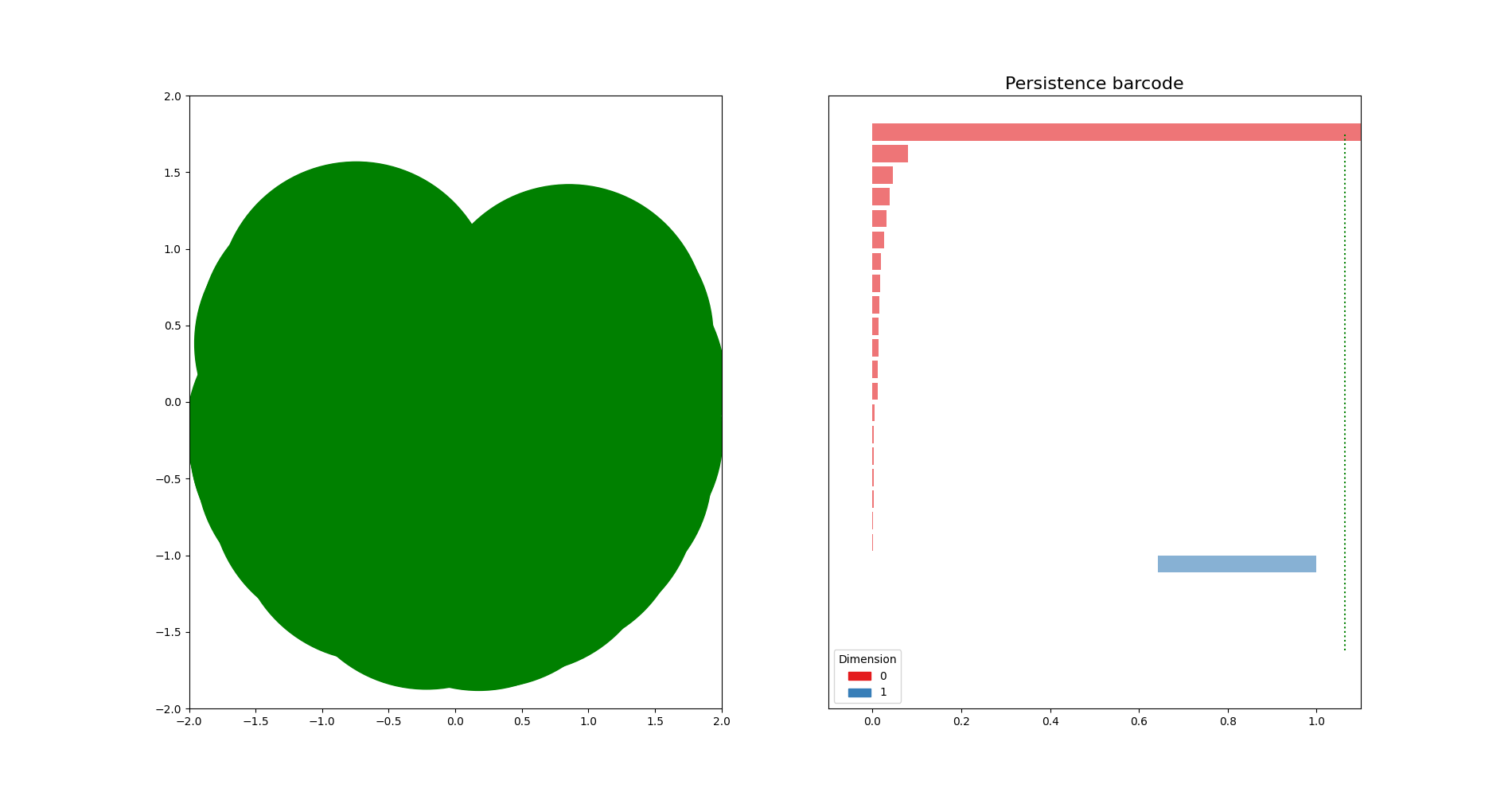
import matplotlib.pyplot as plt
import gudhi as gd
import numpy as np
import math
N=20
np.random.seed(0)
rand_angles = np.random.rand(N) * 2 * math.pi
X = np.stack((np.vectorize(math.cos)(rand_angles), np.vectorize(math.sin)(rand_angles)), axis=1)
fig, axes = plt.subplots(ncols=2, figsize=(4, 2)); fig.set_size_inches(12, 5)
axes[0].scatter(X[:,0],X[:,1], s=50)
axes[0].set_xlim(-2., 2.); axes[0].set_ylim(-2., 2.)
dgm = gd.AlphaComplex(points=X).create_simplex_tree().persistence()
gd.plot_persistence_barcode(dgm, legend=True, axes=axes[1])
plt.show()
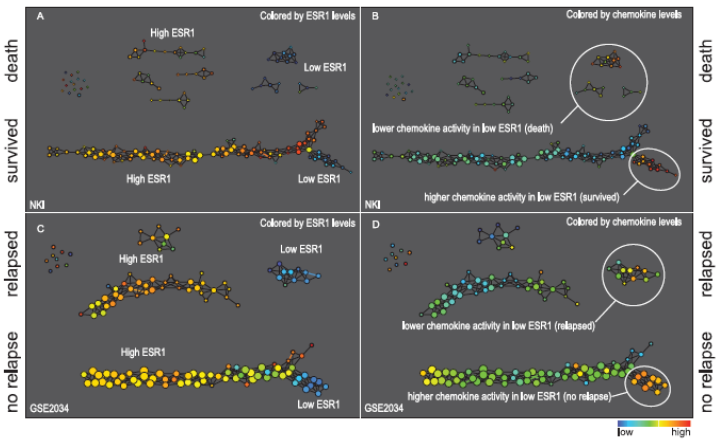
ToMaTo¶
Introduction¶
${\rm ToMaTo}$ algorithm is exposed in the paper of Frédéric CHAZAL et al., Persistence-Based Clustering in Riemannian Manifolds [1], a clustering method based on the idea of topological persistence.
In short, the algorithm needs a density estimation (so to each point $x$ we associate a value $\hat{f}(x)$) and a neighborhood graph. First, it starts with a mode-seeking phase (naive hill-climbing) to build the initial clusters (each with its own mode), following the connected points in the neighborhood graph. Finally, it merges these initial clusters based on their prominence. This merging has a hierarchical nature, i.e. we always obtain the successive new clusters by merging two existing ones.
from sklearn import datasets
import matplotlib.pyplot as plt
import numpy as np
varied = datasets.make_blobs(n_samples=500, cluster_std=[1.0, 2.5, 0.5], random_state=170)[0]
x, y = varied.T
plt.scatter(x,y)
plt.show()
from gudhi.point_cloud.knn import KNearestNeighbors
nbrs = KNearestNeighbors(k=6)
indices = nbrs.fit_transform(varied)
for i in indices:
Y = np.zeros((2,2))
for j in range(len(i)):
Y[0] = [varied[int(i[0])][0], varied[int(i[j])][0]]
Y[1] = [varied[int(i[0])][1], varied[int(i[j])][1]]
plt.plot(Y[0], Y[1], 'ro-')
plt.show()
2. Density¶
from sklearn.neighbors import KernelDensity
kde = KernelDensity(bandwidth=0.1).fit(varied)
z_kde = kde.score_samples(varied)
ax = plt.axes(projection='3d')
_ = ax.scatter(x, y, z_kde, c=z_kde)
plt.show()
from gudhi.clustering.tomato import Tomato
tomato = Tomato()
tomato.fit(varied)
tomato.plot_diagram()
plt.show()
tomato.n_clusters_=3
plt.scatter(x,y,c=tomato.labels_)
plt.show()
%run ./scripts/plot_cluster_comparison.py
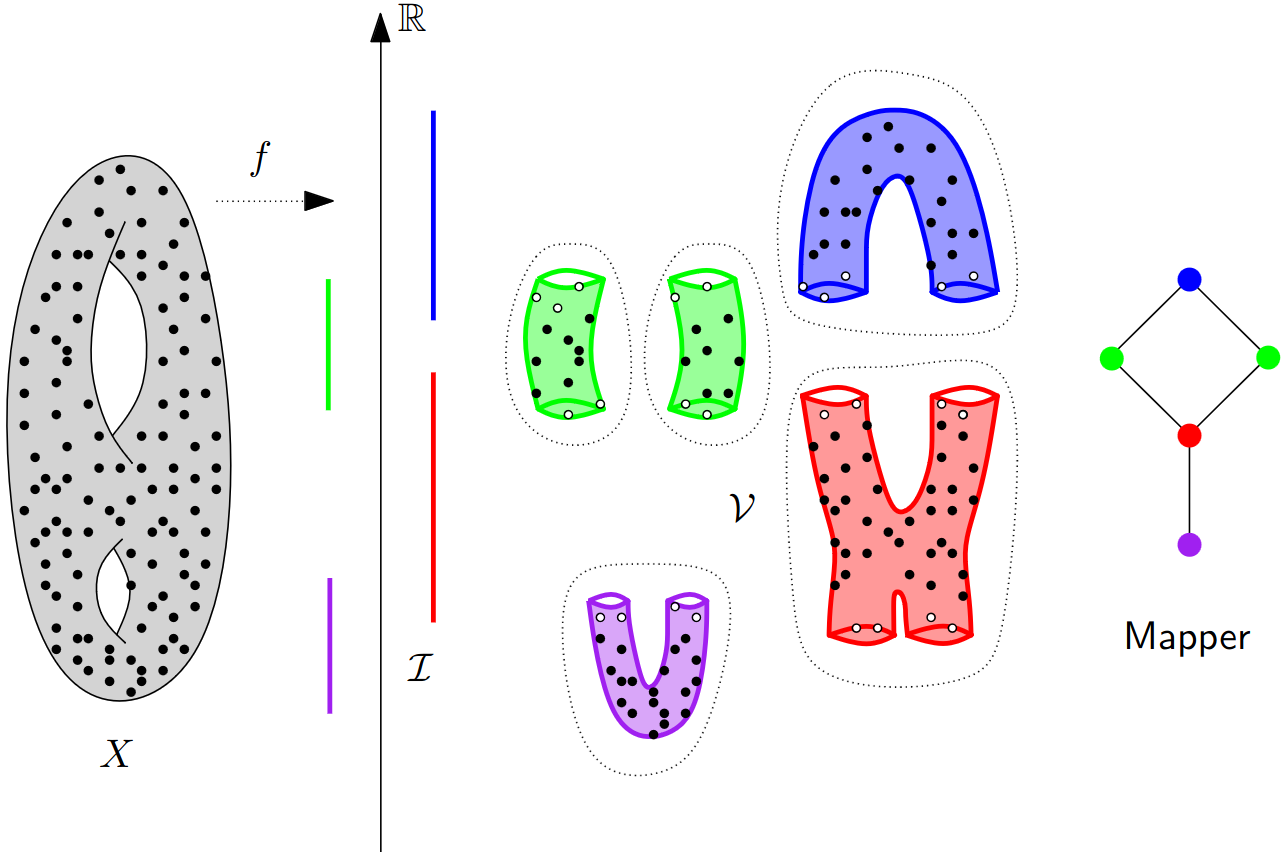
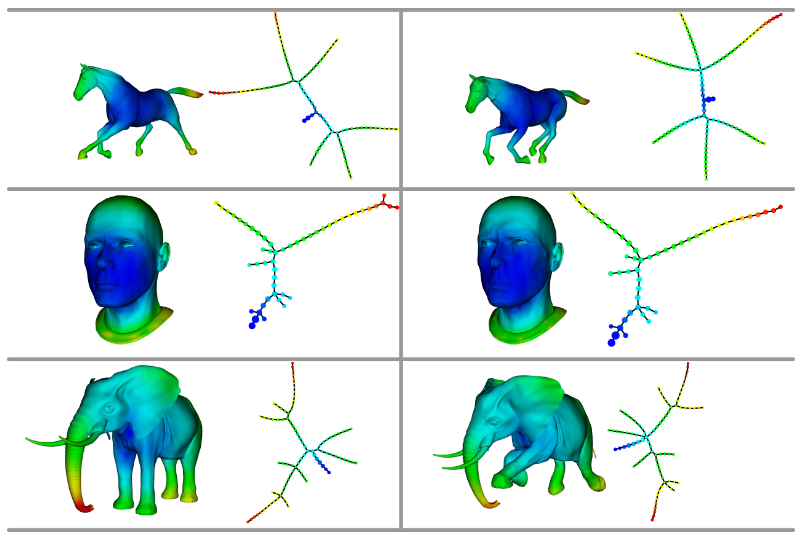
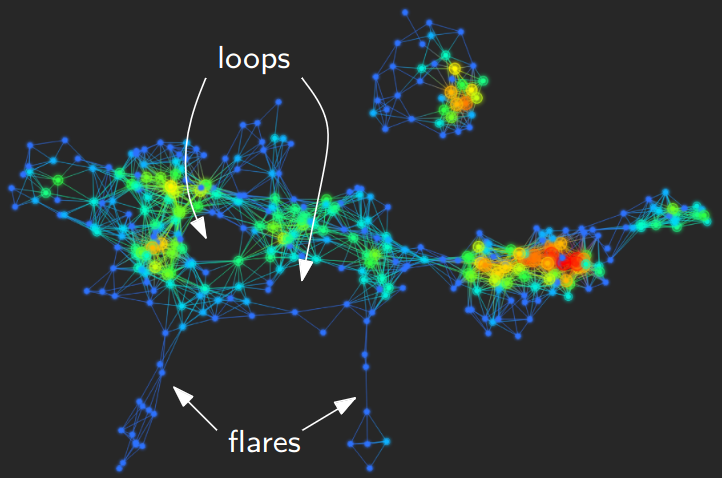
Mapper¶
Visualize topology on the data directly. Two types of applications:
- clustering
- feature selection
Principle: identify statistically relevant sub-populations through patterns (flares, loops)
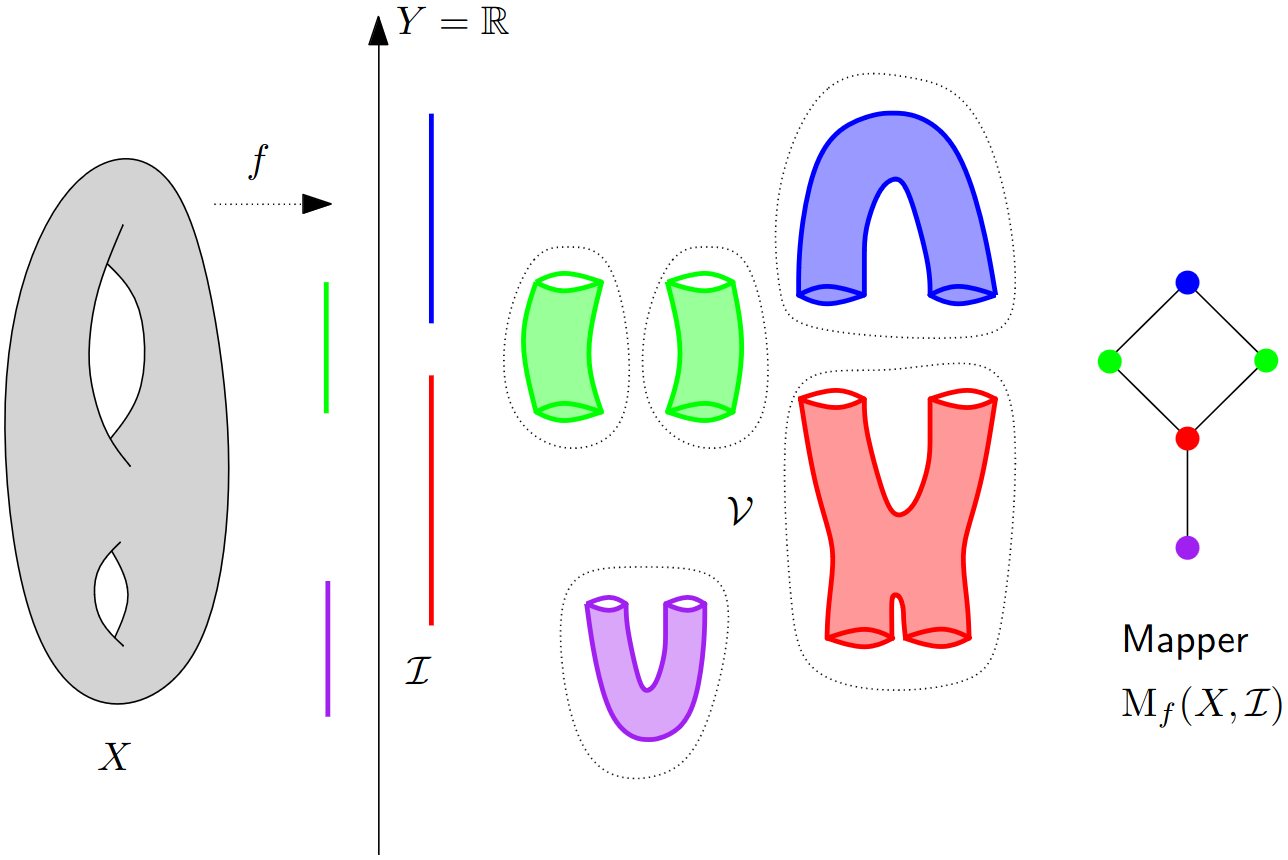
Mapper in the continuous setting¶
Mapper in practice¶